Introduction
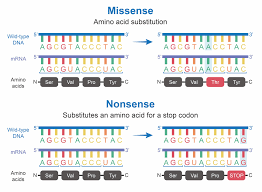
Nonsense Mutation Genetic information is stored in DNA, which serves as the blueprint for building proteins—the essential molecules that carry out nearly all cellular functions. Each gene contains a sequence of nucleotides organized into codons, with each codon specifying a particular amino acid. Protein synthesis relies on this precise sequence, as even a small error can significantly alter the final protein product.
A nonsense mutation is a specific type of point mutation in which a single nucleotide change in DNA transforms a codon that normally codes for an amino acid into a stop codon. The three stop codons in the genetic code—UAA, UAG, and UGA—function as molecular signals for the ribosome to terminate translation. When a nonsense mutation introduces a premature stop codon, the ribosome halts protein synthesis earlier than intended.
The result is a truncated protein, often incomplete and nonfunctional. This interruption can prevent the protein from folding correctly, interacting with other molecules, or performing its biological function. In essence, the genetic instructions are “cut short,” and the resulting protein is unable to fulfill its role in the cell. Depending on the gene affected, nonsense mutations can have serious consequences for cellular function and organismal health, contributing to a range of genetic disorders.
By understanding nonsense mutations, scientists can uncover the mechanisms behind various hereditary diseases and explore therapeutic strategies aimed at mitigating their effects.
How Nonsense Mutations Occur
Nonsense mutations primarily arise from point mutations, specifically substitution mutations, in which a single nucleotide in the DNA sequence is replaced by a different nucleotide. Although this change affects only one “letter” of the genetic code, it can have dramatic consequences for the resulting protein.
For instance, consider the codon UAC, which normally encodes the amino acid Tyrosine. If a substitution occurs that changes this codon to UAA, the new codon becomes a stop codon. During translation, the ribosome interprets this stop codon as a signal to terminate protein synthesis prematurely. As a result, the protein is truncated and frequently nonfunctional.
Even though a nonsense mutation involves only a single nucleotide change, its impact is amplified because it interrupts the reading frame of the gene, producing a protein that may lack essential domains or fail to fold correctly. Depending on the position of the mutation within the gene, the severity of the functional loss can vary—mutations near the beginning of a gene generally produce severely truncated, nonfunctional proteins, whereas mutations near the end may allow partial functionality.
Nonsense mutations can occur spontaneously due to errors in DNA replication or be induced by environmental factors such as radiation or chemical mutagens. Their seemingly minor origin belies the profound biological effects they can exert, making them a critical focus of genetic research and disease studies.

Effects on Proteins and Health
The impact of a nonsense mutation on protein function—and consequently on human health—depends heavily on where the mutation occurs within the gene. Proteins are composed of specific sequences of amino acids that fold into precise three-dimensional structures, allowing them to perform their biological roles. When a nonsense mutation introduces a premature stop codon, this carefully orchestrated process is disrupted.
- Early mutations: If the nonsense mutation occurs near the beginning of a gene, the resulting protein is typically severely truncated. Such proteins are often unstable and recognized by the cell as defective, leading to rapid degradation through quality control mechanisms like the proteasome pathway. This complete loss of functional protein can have catastrophic effects on cellular processes.
- Late mutations: Nonsense mutations occurring toward the end of a gene may yield a partially functional protein. While the truncated protein may retain some activity, it usually operates with reduced efficiency and may be unable to carry out complex interactions or maintain structural stability.
Because proteins are essential for nearly every cellular process—including metabolism, signaling, transport, and structural support—nonsense mutations can result in serious genetic disorders. Diseases arising from such mutations often involve organs or systems where the affected protein plays a critical role, and the severity can range from mild functional impairment to life-threatening conditions.
Understanding the relationship between the location of a nonsense mutation and its biological consequences is crucial for predicting disease severity and for developing targeted therapies to restore protein function.
Examples of Diseases Caused by Nonsense Mutations
Nonsense mutations can have profound effects on human health, as they often lead to nonfunctional or truncated proteins. Several well-characterized genetic disorders are directly linked to these mutations:
- Cystic Fibrosis (CF): Certain forms of cystic fibrosis result from nonsense mutations in the CFTR gene, which encodes a protein responsible for chloride ion transport across cell membranes. When a premature stop codon disrupts the CFTR protein, chloride and water movement is impaired, leading to thick, sticky mucus in the lungs and digestive tract. This contributes to chronic respiratory infections, digestive issues, and reduced life expectancy in affected individuals.
- Duchenne Muscular Dystrophy (DMD): DMD is caused by nonsense mutations in the dystrophin gene, which encodes a protein critical for maintaining muscle fiber integrity. Premature stop codons produce truncated dystrophin proteins that are unable to support muscle cells, leading to progressive muscle weakness, degeneration, and, ultimately, loss of mobility. The severity of the disease is directly linked to the position and nature of the mutation.
- Beta-Thalassemia: This blood disorder arises from nonsense mutations in the HBB gene, which encodes the beta-globin subunit of hemoglobin. When the mutation introduces a premature stop codon, beta-globin production is reduced or halted, causing defective hemoglobin molecules. The resulting anemia can range from mild to severe, leading to fatigue, growth delays, and other health complications.
These examples highlight the critical role that even a single nucleotide change can play in human health. By interrupting the normal coding sequence of genes, nonsense mutations can disrupt essential physiological processes, underlining the importance of genetic research, early diagnosis, and the development of targeted treatments.
Detecting Nonsense Mutations
Accurate detection of nonsense mutations is essential for understanding genetic disorders, guiding treatment decisions, and advancing research. Modern molecular genetics provides a variety of tools that allow scientists and clinicians to identify these mutations with precision.
- DNA Sequencing: This technique determines the exact sequence of nucleotides in a gene, allowing researchers to pinpoint substitutions that result in premature stop codons. By comparing the patient’s DNA sequence with a reference sequence, DNA sequencing can identify the specific location and nature of a nonsense mutation.
- PCR-based Methods (Polymerase Chain Reaction): PCR is used to amplify specific regions of DNA containing a gene of interest. Once amplified, these regions can be analyzed for nucleotide changes that produce stop codons. PCR-based techniques are highly sensitive and cost-effective, making them suitable for targeted mutation screening.
- Next-Generation Sequencing (NGS): NGS represents a high-throughput approach capable of simultaneously analyzing multiple genes or even entire genomes. This technology is especially valuable when the genetic basis of a disease is unclear or when multiple mutations may be contributing to the condition. NGS not only identifies nonsense mutations but also provides a comprehensive view of genetic variation, allowing for more accurate diagnosis and personalized treatment planning.
By combining these methods, scientists can detect nonsense mutations with high accuracy, facilitating early diagnosis, risk assessment, and the development of tailored therapeutic strategies. Detecting these mutations is the first step toward mitigating their potentially severe effects on protein function and human health.
Therapeutic Approaches
Nonsense mutations, by introducing premature stop codons, often lead to nonfunctional proteins and serious genetic disorders. Fortunately, modern medicine has developed several promising strategies to counteract the effects of these mutations and restore protein function.
- Read-through Drugs: These compounds, such as Ataluren, are designed to encourage the ribosome to bypass premature stop codons during translation. By “reading through” the stop signal, the ribosome can continue synthesizing the full-length protein. This approach has shown particular promise in treating genetic disorders like Duchenne Muscular Dystrophy, where restoring even partial protein function can significantly improve patient outcomes.
- Gene Therapy: This approach focuses on directly addressing a faulty gene to repair, replace, or enhance its function. Techniques such as viral vector delivery can introduce a functional copy of the gene into patient cells, allowing them to produce normal proteins. This strategy holds tremendous potential for long-term treatment and even cures for conditions caused by nonsense mutations.
- mRNA Therapy: This innovative approach delivers corrected messenger RNA (mRNA) into cells, enabling them to synthesize functional proteins despite the presence of a defective gene. Unlike traditional gene therapy, mRNA therapy does not permanently alter the genome, reducing some risks associated with gene editing while providing a rapid way to restore protein function.
These therapeutic strategies demonstrate that even severe genetic disruptions caused by nonsense mutations are not necessarily irreversible. Continued research and clinical trials are expanding the range of treatments, offering hope for patients affected by previously untreatable genetic disorders.
Conclusion
Nonsense mutations serve as a striking example of how a single nucleotide change in DNA can have profound biological consequences. By transforming a codon that normally specifies an amino acid into a premature stop signal, these mutations disrupt protein synthesis, often resulting in truncated, nonfunctional proteins. The consequences can be severe, leading to a wide range of genetic disorders that affect essential physiological processes.
Fortunately, advances in molecular genetics and biotechnology have revolutionized our understanding of nonsense mutations. Modern diagnostic techniques allow for precise detection of these mutations, while innovative therapeutic strategies—such as read-through drugs, gene therapy, and mRNA therapy—offer hope for restoring protein function and improving patient outcomes.
Understanding nonsense mutations extends beyond academic interest; it is a critical step in combating hereditary diseases and developing targeted, life-changing treatments. As research continues to evolve, the study of nonsense mutations promises to transform the diagnosis, management, and treatment of some of the most challenging genetic disorders, highlighting the incredible potential of modern science in rewriting the narrative of human health.
FAQs
1. What is a nonsense mutation?
A nonsense mutation is a type of point mutation in DNA where a single nucleotide change converts a codon that codes for an amino acid into a stop codon. This causes the ribosome to halt protein synthesis prematurely, resulting in a truncated and often nonfunctional protein.
2. How is a nonsense mutation different from a missense mutation?
A missense mutation alters a codon so that it codes for a different amino acid, which may affect the protein’s function. In contrast, a nonsense mutation converts a codon into a stop codon, prematurely terminating protein synthesis.
3. Can nonsense mutations occur spontaneously?
Yes. Nonsense mutations can arise spontaneously due to errors during DNA replication or be induced by environmental factors such as radiation, chemicals, or certain viral infections.
4. What are the consequences of a nonsense mutation?
The effects vary based on the location of the mutation within the gene. Early mutations usually produce severely truncated, unstable proteins, while later mutations may produce partially functional proteins. Both can lead to genetic diseases or functional impairments.
5. Which diseases are commonly caused by nonsense mutations?
Some well-known diseases include:
- Cystic Fibrosis (CFTR gene)
- Duchenne Muscular Dystrophy (dystrophin gene)
- Beta-Thalassemia (HBB gene)
6. How are nonsense mutations detected?
They can be detected using techniques such as:
- DNA sequencing (identifies nucleotide changes)
- PCR-based methods (amplifies and screens gene regions)
- Next-Generation Sequencing (NGS) (analyzes multiple genes simultaneously)
7. Are there treatments for disorders caused by nonsense mutations?
Yes. Current therapeutic approaches include:
- Read-through medications (such as Ataluren) enable ribosomes to ignore premature stop codons.
- Using gene therapy to fix or substitute faulty genes.
- mRNA therapy to deliver corrected genetic instructions to cells
8. Can nonsense mutations be inherited?
Yes. If a nonsense mutation occurs in a germline cell (sperm or egg), it can be passed on to offspring. Mutations in somatic cells generally affect only the individual and are not inherited.
9. Why is understanding nonsense mutations important?
Studying nonsense mutations is crucial for diagnosing genetic diseases, developing targeted therapies, and understanding fundamental aspects of protein function. It also helps in advancing personalized medicine and improving patient outcomes.


